Cathepsin D (CD) plays an important role in both biological and pathological processes, although the cleavage characteristics and substrate selection of CD have yet to be fully explored. We employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify the CD cleavage sites in bovine serum albumin (BSA). We found that the hydrophobic residues at P1 were not only a preferential factor for CD cleavage but that the hydrophobicity at P1’ also contributed to CD recognition. The concept of hydrophobic scores of neighbors (HSN) was proposed to describe the hydrophobic microenvironment of CD recognition sites. The survey of CD cleavage characteristics in several proteins suggested that the HSN was a sensitive indicator for judging the favorable sites in peptides for CD cleavage, with HSN values of 0.5–1.0 representing a likely threshold. Ovalbumin (OVA), a protein resistant to CD cleavage in its native state, was easily cleaved by CD after denaturation, and the features of the cleaved peptides were quite similar to those found in BSA, where a higher HSN value indicated greater cleavability. We further conducted two-dimensional gel electrophoresis (2DE) to find more proteins that were insensitive to CD cleavage in CD-knockdown cells. Based on an analysis of secondary and three-dimensional structures, we postulated that intact proteins with a structure consisting of all α-helices would be relatively accessible to CD cleavage.
Cathepsin D as a potential therapeutic target to enhance anticancer drug-induced apoptosis via RNF183-mediated destabilization of Bcl-xL in cancer cells
Cathepsin D (Cat D), a protein predominantly localized in lysosomes, performs multiple biological functions, such as degradation of intracellular and extracellular proteins, regulation of cell death, and activation of inflammatory cells. Its role in cancer cells is worth studying, considering its high expression in several cancers. Cat D increases cancer invasion, metastasis, and angiogenesis , and its overexpression increases the risk of recurrence and death in female patients with breast cancer. In addition, Cat D is required for migration and invasion of gastric and breast cancer cells. However, its effect on cell death is controversial and depends on stimulators and cell context. Cat D protects cancer cells from acetate- and oxidative stress-induced cell death. In contrast, it potentiates cell death in anticancer drug-treated cells . The role of Cat D has been well-known in breast cancer. Pro-Cat D is released from triple-negative breast cancer, and increases proliferation of cancer cells . Extracellular Cat D induces proteolysis of SPARC C-terminal extracellular Ca2+ binding domain, leading to stimulation of migration and invasion in triple-negative breast cancer . Cat D-targeted antibody inhibits tumor growth in triple-negative breast cancer . Recently, the role of intracellular Cat D, but not of its secreted form, has been identified. In PyMT cells, Cat D deficiency delays tumor cell proliferation through inhibition of mTORC1 signaling .
Cathepsins
Cathepsins are a class of lysosomal proteases that play important roles in proteolysis during physiological processes. They are reportedly involved in a number of diseases, such as cancer, atherosclerosis, arthritis and neurodegenerative diseases. Several cathepsins can function outside of cells. For example, cathepsins B, D and L are able to cleave proteins in the extracellular matrix (ECM), including collagen , fibronectin , proteoglycans and laminin , and are considered to represent causal factors in tumor invasion and metastasis. Therefore, identification of the native substrates and cleavage sites of cathepsins is necessary to understand their physiological and pathological roles.
Cathepsin D (CD) is an aspartic endoprotease that is widely distributed in mammalian cells. Because of dual locations of CD, being either located in organelles such as in the cytoplasm, lysosomes and phagosomes, or secreted into the ECM, it participates in a number of physiological processes, including cell proliferation , apoptosis , senescence and tissue homeostasis . CD is also known to take part in various pathological processes; it is likely involved in cancer development as well as metastasis , atherosclerosis and Alzheimer’s disease . Like other cathepsins, CD recognizes its substrate with a relatively low selectivity. Nevertheless, it does not function on some proteins under certain circumstances, such as hen egg white lysozyme and ovalbumin (OVA).
Previous investigations of CD cleavage sites were initiated using synthetic peptides and medium-sized natural peptides, which revealed that the amino acid residues at P2, P2’, P3 and P3’ exerted some influence on the susceptibility to CD cleavage and the hydrophobic residues co-occupying P1 and P1’ favored CD attack .
Fluorogenic Substrates for Cathepsin D
Fluorogenic substrates for cathepsin D; A-Tyr-Phe(NO2)-Leu-Leu (A; Ala-Arg-Pro-Lys-Pro-Leu-Leu-, Arg-Pro-Lys-Pro-Leu-Leu-, Pro-Lys-Pro-Leu-Leu-, Lys-Pro-Leu-Leu-, Pro-Leu-Leu-) and B-Phe(NO2)-Tyr-Leu-Leu (B; Arg-Pro-Lys-Pro-Leu-Leu-, Pro-Lys-Pro-Leu-Leu-, Lys-Pro-Leu-Leu-, Pro-Leu-Leu-) (Phe(NO2), p-nitrophenylalanine) were synthesized and digested by cathepsin D and pepsin. The fluorescence at 303 nm (excitation at 260 nm) was increased with the hydrolysis of the substrates. The minimum detectable cathepsin D concentrations for these substrates were 0.5-4 nM and pepsin concentrations were 0.1-0.8 nM except Pro-Leu-Leu-Tyr-Phe(NO2)-Leu-Leu under the following conditions: substrate concentration, 20 μM; measuring time, 3 min. The hydrolysis rate constants (kcat/Km) of B-Phe(NO2)-Tyr-Leu-Leu for cathepsin D were same or 2-3 times greater than A-Tyr-Phe(NO2)-Leu-Leu. On the other hand, those of B-Phe(NO2)-Tyr-Leu-Leu for pepsin were the same or 4-20 times greater than A-Tyr-Phe(NO2)-Leu-Leu. The hydrolysis rates of the substrates by both enzymes tend to increase with the increase of the peptide chain length. The best substrate for cathepsin D was Arg-Pro-Lys-Pro-Leu-Leu-Phe(NO2)-Tyr-Leu-Leu and its kcat/Km was 1.3 μM-1 s-1.
Proteins Cleaved by CD
As the spatial structures of proteins are different from peptides, the information on cleavage sites favored by CD derived only from peptides is not sufficiently convincing to demonstrate the targeted sites in proteins. The CD cleavage sites in some native proteins, such as bovine serum albumin (BSA), hemoglobin , actin , antichymotrypsin and kallistatin, were therefore examined individually and hydrophobic amino acids potentially associated with the scissile bonds were proposed as targeted sites. Moreover, it was noted that CD preferred sites involved in the α-helical conformation of myoglobin and cytochrome c . Recently, proteomic approaches have emerged as a powerful tool for screening the protein substrates and characterizing the features of proteases. Global profiles have indicated that CD proteolysis mainly occurs between hydrophobic residues, with a strong preference for leucine and phenylalanine . Additionally, the cleavage activity of CD in target proteins not only relies on the linear sequences of amino acids but is also decided, at least in part, by the protein’s spatial structure.
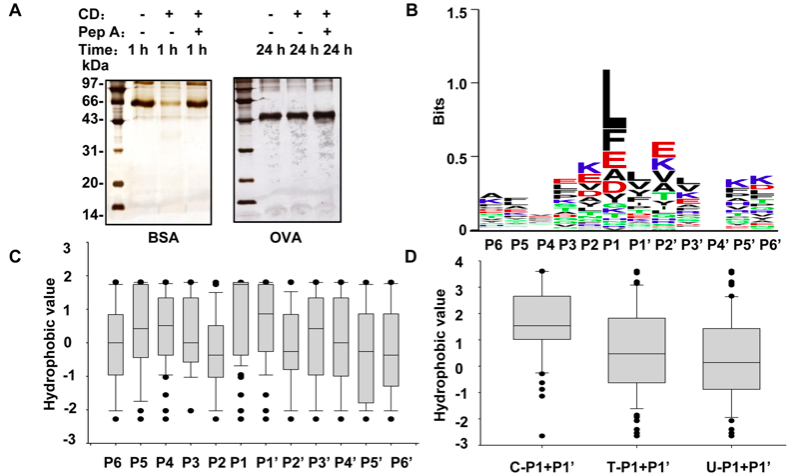
In order to assess the favorable conditions of CD cleavage, the proteins that are sensitive or insensitive to CD cleavage like BSA and OVA, were incubated with CD under different digestion conditions, such as buffer pHs, reaction temperatures and incubation time. After evaluating all the conditions of CD cleavage, especially considering to detect the amino acid sites more susceptible to CD cleavage, optimal reaction conditions were selected in which cleavage targets at a final concentration of 1.5 µM were incubated with 0.2 U CD in 100 mM sodium citrate buffer (pH 3.5) for 3 h at 37°C. To estimate the efficiency of CD cleavage, the mixtures of the targets and CD before and after incubation were loaded onto 12% SDS-PAGE gels, followed by silver nitrate staining.
There is no parameter selection for CD digestion in protein search engines such as Mascot and SEQUEST. Therefore, setting no-enzyme in Mascot was used to search for peptides generated by CD based on MS/MS signals. 1) there are no conservative motifs among the BSA peptides; 2) there is no obvious preference for amino acid residues at positions other than P1; 3) several residues are much more preferred by CD at P1, such as L, F, E, A and D, which are all hydrophobic amino acids (the acidic amino acids become a little hydrophobic at pH 3.5), and 4) although P1 is dominantly occupied by hydrophobic residues with a total occurrence frequency nearly 83%, 7 kinds of hydrophilic amino acids appear at P1 with a total of 17% cleavage preference. The occurrence frequencies of the residues at each position in the logos are listed in Table S3. These results are generally in agreement with previous observations that the hydrophobic residues are favored by CD.
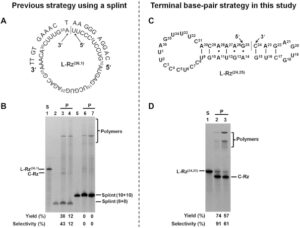
Considering that CD cleavage is likely correlated with the hydrophobic microenvironment contributed by P1 and P1’, we introduce a new concept to describe the hydrophobicity of neighboring residues referred to as the hydrophobic scores of neighbors (HSN), where an HSN value represents the sum of the hydrophobic scores of two neighboring residues. We analyzed the CD cleavage characteristic of BSA in term of HSN. As approximately 81% of the P1 positions in the CD-cleaved BSA peptides were occupied by the 6 residues: L, F, E, A, D and Y, we extracted all the P1/P1’ residue pairs in BSA sequence that consist of all the 6 residues (as P1) and their corresponding neighbor residues (as P1’), and broadly divided these pairs into 3 groups: all of the P1/P1’ pairs in BSA sequence (T); all of the pairs detected via LC-MS/MS (C); and all of the pairs undetected (U). The HSN value for a P1/P1’ pair is the sum of the hydrophobic sores of the residue at P1 and its neighboring residue at P1’, which was calculated according to Cowan method . Figure 1D presents the median HSN values from each group and demonstrates that the median value in the C group is significantly higher than those of the other two groups. Moreover, the HSN occurrence frequency, denoted as the ratio of the number of the P1/P1’ pairs with certain HSNs to the total pair number, was plotted against the scale of the HSN intervals to determine which HSN value could be regarded as the threshold for CD cleavage of BSA. The data presented in Figure 1E demonstrate that the HSN interval of 0.5–1.0 serves as a cutoff, with all of the HSN occurrence frequencies in the C group higher than U falling on the right, while those in the U group greater than C fall on the left. Our results therefore suggest that an HSN value of 0.5–1.0 in BSA is an indicator of CD cleavage.

If protein secondary structure does play a key role in CD access, the coil structure between α-helix and β-sheet is likely a considerable factor to regulate such susceptibility. On the basis of the currently available data, we could not draw a conclusion that how to set up a structural threshold for CD access to a protein. On the other hand, two deductions are acceptable, 1) a protein with all α-helices is sensitive to CD cleavage, whereas a protein with all β-sheets is insensitive to CD; and 2) once the spatial structure of a protein insensitive to CD cleavage is cracked, CD can attack its scissile sites with high HSN values.